DEEP DEVICE MINING
Search Functions For Enzyme Discovery and Pathway Engineering
The microcosmos provides an immense reservoir of genomic diversity in the form of functional genes and pathways that can be used in biological engineering applications. We apply conventional substrate conversion and biosensor-based high-throughput screening methods (i.e. deep device mining) for the recovery of biocatalysts from natural and engineered environments.
Ecosystem Type: Environmental and host-associated sources
Location: Variable with emphasis on Canadian sampling sites
The Art and Design of Functional Metagenomic Screens
An increasingly important tool in biological engineering is deep device mining based on recovering genomic information (DNA) from the environment encoding activities of interest that can in turn be used to reprogram industrial strains or microbial communities. Engineered microbial communities expand upon the standardized part concept to include cellular organisms as carbon-based information processing units implementing a distributed metabolic algorithm. These BioFactories perform complex tasks more effectively than single cells, complement enzyme complexes mediating biomass conversion, and resist environmental perturbation.
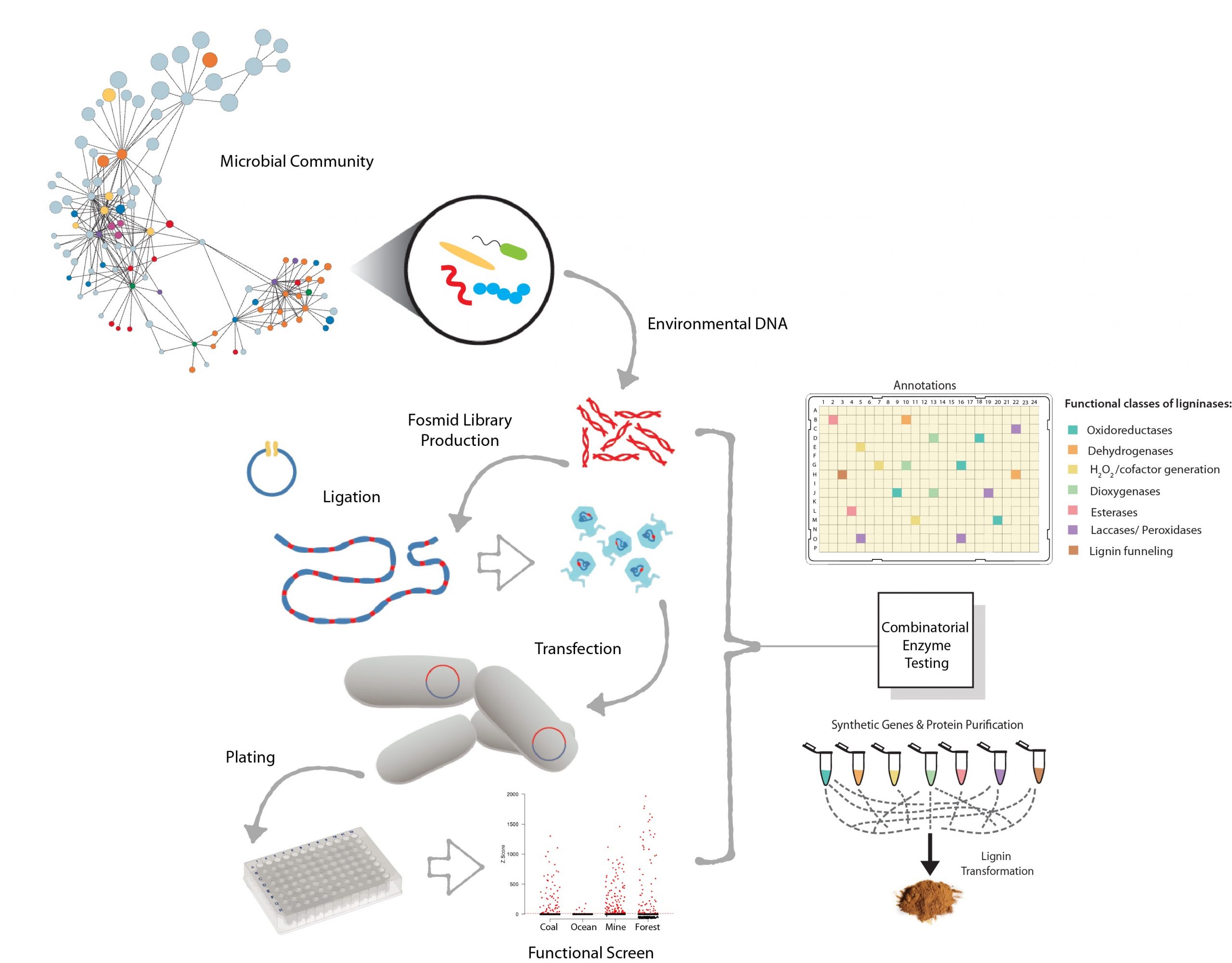
We have developed a functional screening platform leveraging automation systems in the Biofactorial High-throughput Facility, involving large-insert (32-47kb) metagenomic (fosmid) libraries hosted in E.coli supporting high-throughput recovery and identification of biocatalysts from uncultivated microbial communities (PMID: 26231123, PMID: 21440432, PMID: 21307835, and PMID: 19776717). This screening platform, in combination with labeled substrates and whole cell biosensing, has successfully identified carbohydrate active enzymes (CAZymes) and putative ligninases from diverse phylogenetic backgrounds capable of catalyzing cellulose and lignin transformation. More recently we have developed selection strategies to enrich fosmids encoding resistance phenotypes to a wide range of lignin-derived monoaromatic compounds. Relevant primary publications related to functional screening include (PMID: 31182795, PMID: 31164449, PMID: 31080075, PMID: 30013164 and PMID: 23906845).
Figure 1. A workflow for constructing large insert (fosmid) libraries. Library production involves high molecular weight environmental DNA preparation, ligation into a vector backbone and “head-full packaging” of ligated DNA into a phage delivery system. Host cells are then transfected, plated and arrayed in 384-well plate libraries which can be searched using different screening paradigms such as biosensors or substrate conversion. Fosmid clones resulting in a signal above a significance threshold are selected for sequencing and functional characterization. Positive clones identified in functional screens can be sequenced and the gene or gene cluster conferring functional activity identified using a combination of in silico and experimental methods. Subsequent annotations are categorized into functional classes relevant to lignin transformation and used to select specific genes for synthesis and protein expression. Purified proteins are tested in a combinatorial array to identify synergistic interactions driving enhanced lignin transformation.
Biosensors as a Search Function
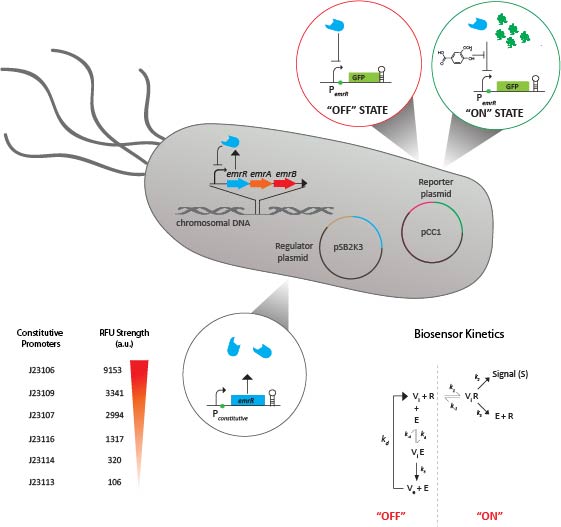
Microbes have evolved myriad genetic circuits used to sense and respond to small molecules in both intra- and extracellular environments including cell surface receptors, transcription factors, and RNA aptamers. We refactor allosteric transcription factors and cognate promoters into standardized whole-cell biosensors that produce a fluorescence signal in response to specific small molecule interactions. The application of whole-cell biosensors in high-throughput functional screens allows us to search for biocatalysts in large insert genomic libraries in co-culture. We have applied this approach to develop a search function in which a transcriptional regulator produces a fluorescent signal in response to vanillin and syringaldehyde (specific monoaromatic products of lignin transformation) and continue to develop new sensors for lignocellulosic and plastic polymer transformation. Relevant primary publications related to biosensor development include (PMID: 24982175 and PMID: 29182267 )
We are actively exploring the use of droplet methods to expand our screening search space with reduced cost and carbon footprint. This new screening paradigm involves using oil encapsulated droplets that are created and sorted in microfluidic chips. Instead of screening hundreds of large insert DNA clones in microtiter plates, we can screen millions of droplets at a time, enabling new economies of scale in deep device mining. The platform is compatible with any fluorescence-based assay, including fluorogenic substrates and biosensors. In addition to sorting, we can store hundreds of thousands of droplets in custom storage chips, enabling measurements of enzyme kinetics on a cell-by-cell basis.”
Figure 2. PemrR-GFP biosensor for detection of lignin transformation products, syringaldehyde and vanillin. The promoter-operator sequence of the emrRAB operon is inserted upstream of GFP on a pET15 plasmid. The transcriptional regulator EmrR is expressed endogenously by E. coli, and binds to the promoter-operator sequence of emrRAB. The binding of EmrR to the promoter-operator sequence of the PemrR-GFP biosensor represses transcription of GFP (‘OFF state’). Upon induction by monoaromatic compounds such as vanillin and syrinaldehyde, the EmrR transcriptional regulator is de-repressed, which initiates GFP (‘ON state’).