GENOME ENGINEERING
We develop molecular tools for the rational modification of microbial genomes. By combining modern and classical techniques, including CRISPR, recombination and transposition systems, as well as developing new vector systems, we are able to more effectively study and engineer microorganisms for basic and applied research outcomes at the individual, population and community levels of biological organization.
F– λ– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 Δ(lac)X74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK rpsL (StrR) nupG’ trfA dhfr
Molecular Tools and Modular Toolkits
Microorganisms have evolved to solve complex metabolic problems over approximately 4 billion years of evolution, innovating distributed solutions to nutrient and energy conversion processes that have fundamentally transformed the surface chemistry of the earth and generated a deep reservoir of genetic information integral to the emergence of multicellular life including our own through successive symbiotic innovations. The concept of distributed metabolism has emerged as a topic of interest in synthetic biology and biological engineering. Synthetic biology designs, builds, and tests biological or biologically-inspired systems using standardized parts composed of DNA that conform to an assembly standard. Scientists engineer cellular “chassis” such as Escherichia coli by combining standard parts into integrated biological devices and learn from resulting outputs. We are developing and applying various genome engineering strategies with applications in high-throughput screening, metabolic engineering and novel gene discovery in both E. coli and cyanobacterial chassis.
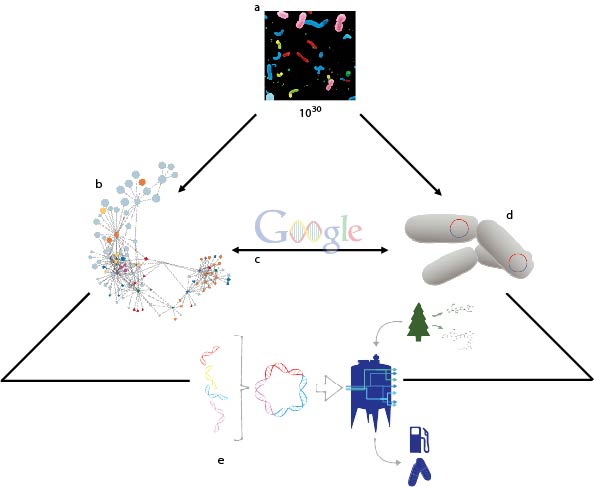
Figure 1. Harnessing the hidden metabolic power of the microcosmos. (a) The invisible microbial majority drives energy and material transformations through (b) distributed networks of metabolite exchange that can be (c) interrogated with biological search functions that identify metabolic processes that can be (d) programmed into cellular chassis or synthetic microbial consortia (e) that are in turn used to develop sustainable modes of material and energy production.
Specific tool development includes the application of CRISPR-Cas9 methods for high-throughput identification of active genes or gene cassettes discovered in screening and whole-genome knock-out studies of cellular networks in cyanobacteria. In parallel, we are adapting chassis-independent recombinase-assisted genome engineering (CRAGE) for stable integration of gene cassettes and clusters into both E. coli and cyanobacterial chromosomes. We apply these tools to improve the sensitivity and throughput of functional screening workflows and to engineer pathways based on screening outcomes. CRISPR-interference enables us to quickly investigate gene function of active screening hits. By targeting endogenous chromosomal genes, we can also redirect energy and cellular resources towards specific metabolic pathways and processes and test genetic engineering strategies. Use of the CRAGE system allows us to both monitor population dynamics in co-culture-based screens and to insert new functions into specific host backgrounds iterating rapidly in the design, build, test, learn cycle.
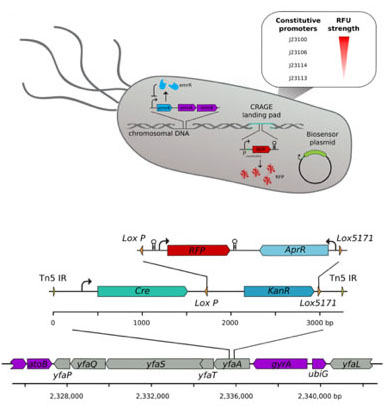
Figure 2. CRAGE-enabled engineering of E. coli EPI300 reporter strains. The EmrR transcriptional regulator enables monoaromatic-dependant GFP expression following transformation with the pSB1C3-PemrR-GFP biosensor plasmid. Four constitutive RFP expression cassettes, with a range of promoter strengths, were integrated into the chromosome as a method of labelling the reporter strain (top). Chromosomal integration was performed using the CRAGE system, requiring landing pad transposition into the EPI300 chromosome. This enables high-efficiency Cre-lox-recombination into the CRAGE landing pad site located in the nonessential yfaA locus in this example (bottom).
