SYNTHETIC MICROBIAL CONSORTIA
Distributing Metabolic Functions
We are using selection-based screening approaches to recover a minimum set of environmental genes and pathways encoded in fosmid library pools that are needed to implement a specific metabolic function. This allows us to discover and characterize single genes as well as synergistic interactions between different clones enabling distributed metabolic processes at the population and community levels of biological organization.
Ecosystem Type: Environmental and host-associated sources pooled into isogenic populations harboring diverse metagenomic clones
Fosmid Pool Selection
We use fosmid library pools as an experimental and systems-level model for recovering enzymatic networks that enable distributed metabolism and cooperativity between fosmid clones. Fosmid library pools act as synthetic microbial communities programmed into a uniform genetic background that can be shaped through selection to solve specific metabolic problems in an unsupervised manner. We are currently focused on recovering enzymatic networks that enable distributed metabolism and cooperativity between fosmid clones involved in lignin transformation.
We hypothesize that lignin transformation is a multi-locus phenotype distributed between different members of a community and requires multiple pathway components from initial depolymerization to biofuneling of lignin oligomers and monomers. Therefore, pooled fosmid libraries represent a novel and promising approach for reconstructing lignin transformation phenotypes sourced from environments where lignin transformation activities are prevalent. Our pooling strategies will enable us to recover synergistic interactions between several environmental genes enabling combinatorial lignin transformation in multi-step processes. We have demonstrated the feasibility of this approach in a previous study on combinatorial lignin transformation from coal bed fosmid clones (PMID: 24982175).
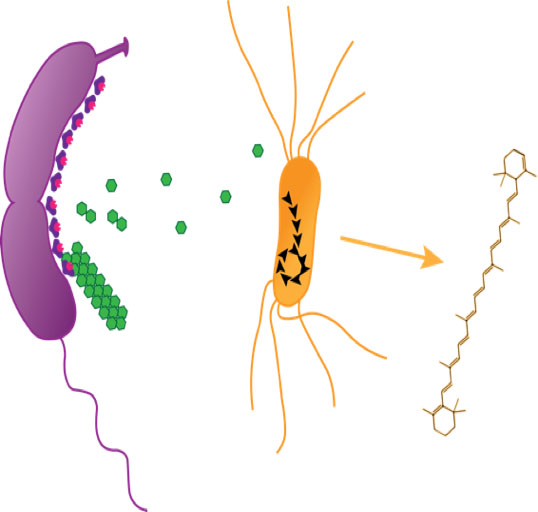
Figure 1. A workflow for pooling fosmid libraries and application of genetic information derived from pool selection or enrichment. Library pools are selected or enriched in LB media containing lignin as a source carbon source with or without glucose priming. Resulting clones are sequenced to completion and analyzed in silico. Selected genes of interest are synthesized and cloned into C. crescentus for S-layer cell-surface display. Engineered C. crescentus can be combined with different S-layer arrays or other bacterial strains to construct synthetic microbial consortia optimized for consolidated bioprocessing.
Figure 2. Whole cell biocatalysts engineering. To eliminate the necessity to purify lignocellulolytic enzymes to breakdown insoluble biomass polymers, we are developing a robust enzyme display platform on the cell surface of Caulobacter crescentus that can be used to funnel metabolites into E. coli metabolism for the production of value-added compounds such as terpenoids.